jumble: Random assignment
Kevin W. McConeghy
2020-01-06
Source:vignettes/03-assignment.Rmd
03-assignment.RmdWelcome to jumble! This program was written as a companion to academic work performed by researchers at Brown University to conduct re-randomization for cluster-randomized nursing home trials.
Introduction
In this vignette we demonstrate some functions used for random assignment, following best practices outlined by other R packages like randomizeR, randomizr etc.
- Randomization must be transparent
- Randomization must be reproducibile
- Randomization must be random!
A note on random number generation
The random sampling method underlying jumble functions is the base R function sample(), rbinom(). While computational speed may be problematic in some cases, the base sample function is reliable, stable, and well-documented and tested.
The sample() functions calls the default Random number generator for R, the Mershene-Twister method, which can be queried using RNGKind().@Mersenne1998
Current system build:
## R version 3.6.1 (2019-07-05)## Windows platform : x86_64-w64-mingw32/x64 (64-bit)Random assignment to K-groups
Assigning treatment is easy if you want equal probability of assignment to one or more groups, without concern to final sample size in each arm, covariate balance etc.
## # A tibble: 6 x 2
## id group
## <int> <chr>
## 1 10124 a
## 2 10140 b
## 3 10165 b
## 4 10190 a
## 5 10229 a
## 6 10361 aTruly random assignment!
The seed is stored as a attribute
## [1] "Randomization performed: 2020-01-06 Seed: 18073"Don’t lose that information! As long as you know the seed you can reproduce the randomization:
## # A tibble: 6 x 2
## id group
## <int> <chr>
## 1 10124 a
## 2 10140 b
## 3 10165 b
## 4 10190 a
## 5 10229 a
## 6 10361 aSimulation to demonstrate equal probability
Perform 20,000 randomizations
pidnum <- df$pidnum
n=20000L
df_assign <- lapply(1:n, function(x) rnd_assign(pidnum, 2, 18073L+x))
assigns <- bind_cols(df_assign[[1]][, 1],
lapply(df_assign, function(x) x[, 2]))
df_assign_2 <- gather(assigns, key = 'trial', value = 'group',
starts_with('group')) %>%
group_by(id) %>%
summarize(`No. of trials` = n,
`No. group 'a'` = sum(group == 'a'),
`No. group 'b'` = sum(group == 'b'),
`Prob. group = 'a'` = `No. group 'a'` / n(),
`Prob. group = 'b'` = `No. group 'b'` / n())
head(df_assign_2)## # A tibble: 6 x 6
## id `No. of trials` `No. group 'a'` `No. group 'b'` `Prob. group = …
## <int> <int> <int> <int> <dbl>
## 1 10124 20000 9986 10014 0.499
## 2 10140 20000 10000 10000 0.5
## 3 10165 20000 9997 10003 0.500
## 4 10190 20000 10003 9997 0.500
## 5 10229 20000 9997 10003 0.500
## 6 10361 20000 10001 9999 0.500
## # … with 1 more variable: `Prob. group = 'b'` <dbl>First glance shows ~ equal likelihood of group assignment in 20,000 trials. ### Assignment tests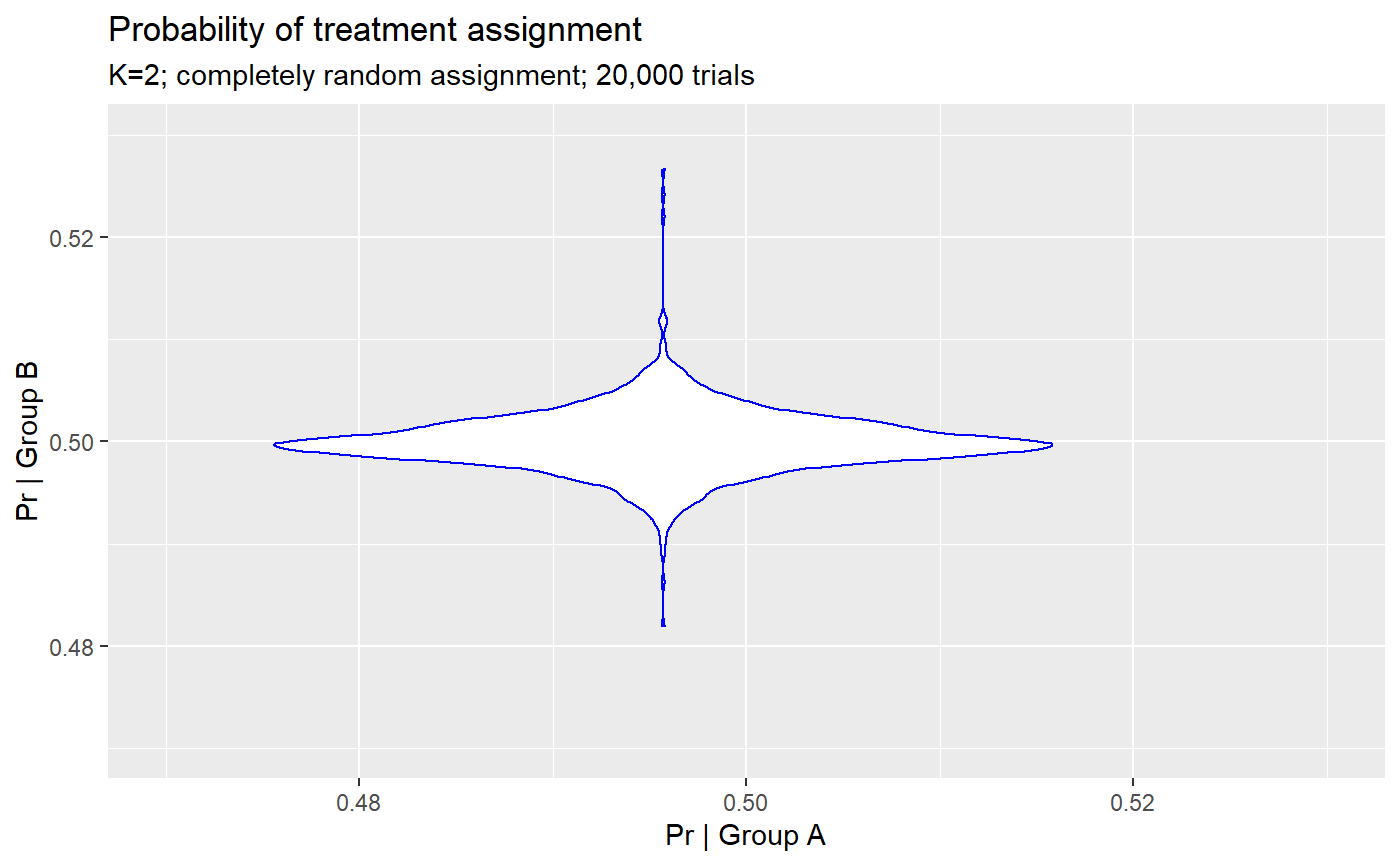
For any given individual, across 20,000 trials no assignment exceeds an imbalance of ~2-3%.
## Probability of assignment to group 'a' in 20,000 trials## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.4733 0.4984 0.5001 0.5000 0.5015 0.5181## Probability of assignment to group 'b' in 20,000 trials## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.4819 0.4985 0.4999 0.5000 0.5016 0.5266However, in each trial the number of individuals in group A vs. B was random and could be imbalanced.
df_trials <- gather(assigns, key = 'trial', value = 'group',
starts_with('group')) %>%
group_by(trial) %>%
summarize(`No. group 'a'` = sum(group == 'a'),
`No. group 'b'` = sum(group == 'b'))## No. assigned to group 'a' in 20,000 trials## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 459.0 516.0 527.0 526.9 538.0 588.0## No assigned to group 'b' in 20,000 trials## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 466.0 516.0 527.0 527.1 538.0 595.0See how the number assigned varies widely?
what if you want to guarantee an equal number of individuals are in each group?
Random assignment to K-groups, equal sample size
We will repeat the exercise, dropping one person to make an odd-numbered cohort.
pidnum <- df$pidnum
pidnum <- pidnum[2:length(pidnum)] # drop 1
n=10000
seeds <- sample(-100000:100000, n)
df_assign <- lapply(1:n, function(x) {
set.seed(seeds[x])
rnd_allot(pidnum)
})
assigns <- bind_cols(df_assign[[1]][, 1],
lapply(df_assign, function(x) x[, 2]))
df_assign_2 <- gather(assigns, key = 'trial', value = 'group',
starts_with('group')) %>%
group_by(id) %>%
summarize(`No. of trials` = n,
`No. group 'a'` = sum(group == 'a'),
`No. group 'b'` = sum(group == 'b'),
`Prob. group = 'a'` = `No. group 'a'` / n(),
`Prob. group = 'b'` = `No. group 'b'` / n())
head(df_assign_2)## # A tibble: 6 x 6
## id `No. of trials` `No. group 'a'` `No. group 'b'` `Prob. group = …
## <int> <dbl> <int> <int> <dbl>
## 1 10140 10000 4958 5042 0.496
## 2 10165 10000 4955 5045 0.496
## 3 10190 10000 5026 4974 0.503
## 4 10229 10000 4959 5041 0.496
## 5 10361 10000 4971 5029 0.497
## 6 10368 10000 4943 5057 0.494
## # … with 1 more variable: `Prob. group = 'b'` <dbl>Assignment tests
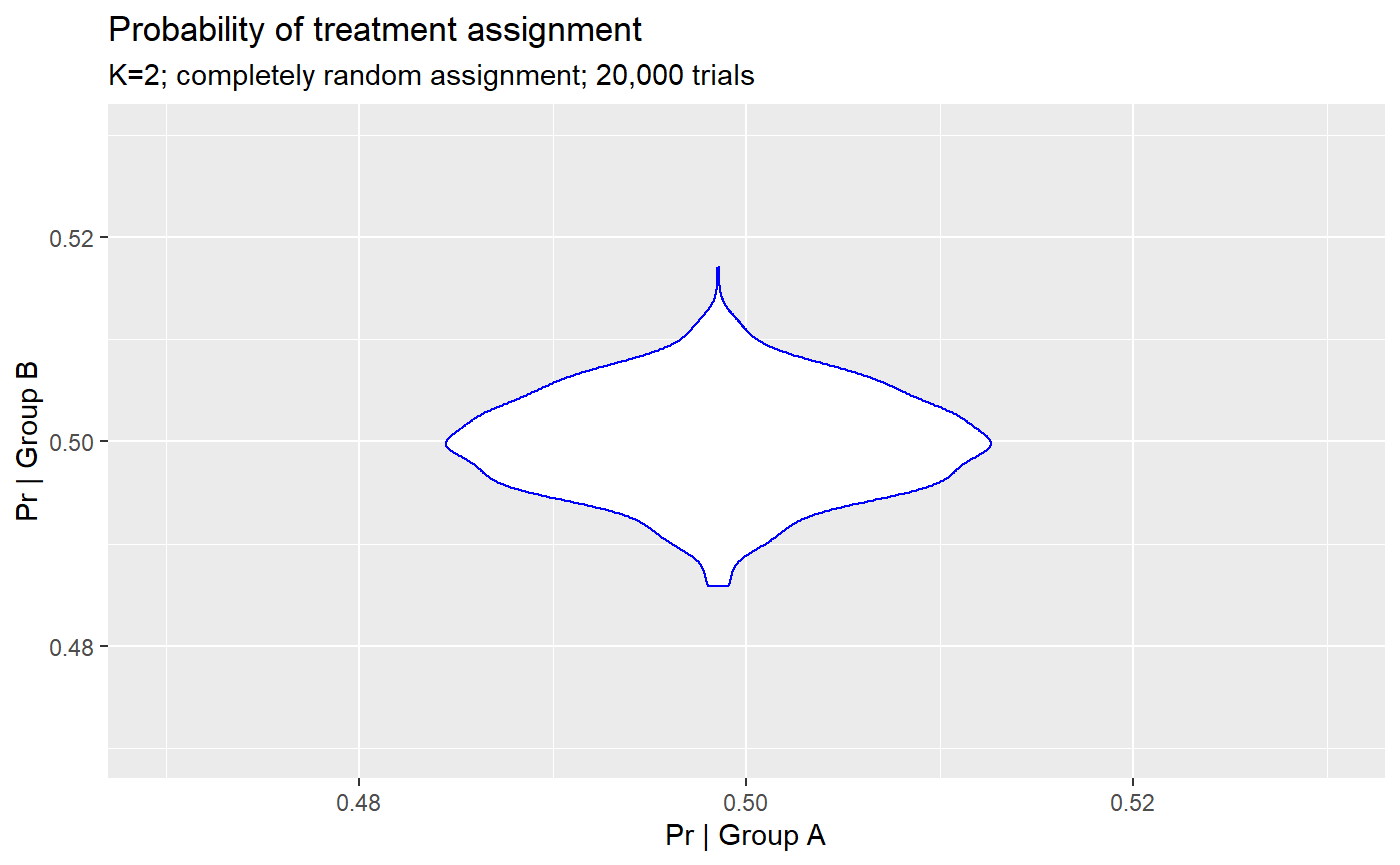
Random assignment looks well-balanced between groups.
## Probability of assignment to group 'a' in 20,000 trials## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.4829 0.4965 0.5001 0.5000 0.5035 0.5142## Probability of assignment to group 'b' in 20,000 trials## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.4858 0.4965 0.4999 0.5000 0.5035 0.5171df_trials <- gather(assigns, key = 'trial', value = 'group',
starts_with('group')) %>%
group_by(trial) %>%
summarize(`No. group 'a'` = sum(group == 'a'),
`No. group 'b'` = sum(group == 'b'))What about the size of the groups?
## No. assigned to group 'a' in 20,000 trials## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 526.0 526.0 526.0 526.5 527.0 527.0## No assigned to group 'b' in 20,000 trials## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 526.0 526.0 527.0 526.5 527.0 527.0Equal size! The number now randomly goes between 526 or 527 for either the ‘a’ or ‘b’ group. The imbalance is selected with equal probability.